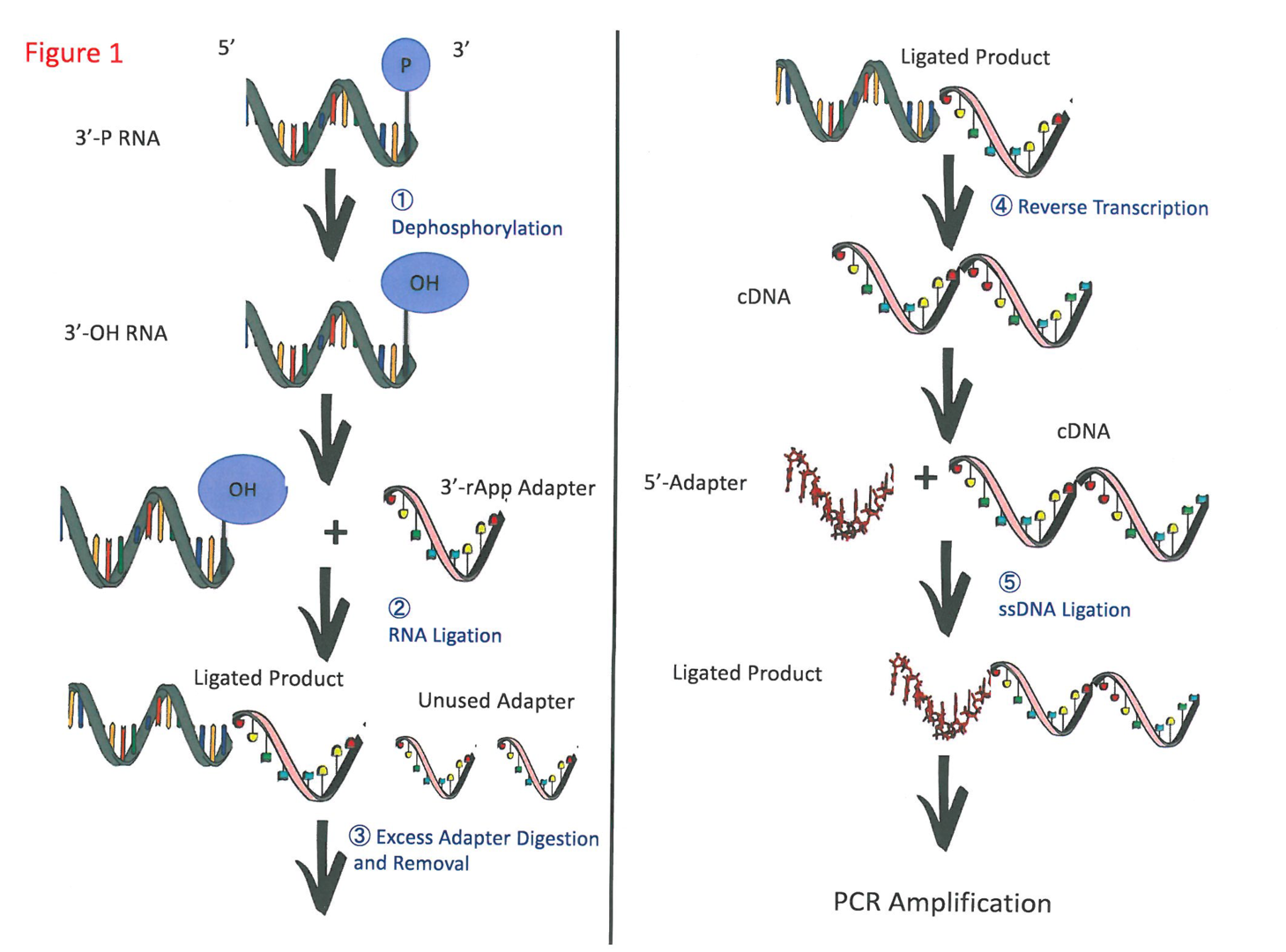
Opportunity
This invention provides a new and important platform for constructing cDNA libraries for a wide range of biological applications. Using the strategy employed here can likely produce a cDNA library with high yield, in significantly reduced time and at a reasonably low cost, while having less side-product/contamination than current technologies. Notably, the sensitivity of this method will enable low-input RNA samples (e.g., those from rare cells and tissues) to be used for cDNA library preparation. Not only can this invention be used in RNA structurome studies, but also in other RNA-related, next-generation sequencing (NGS) applications such as RNA-sequencing (RNA-seq). As the global NGS-based RNA-seq market is projected to reach 2.65 billion USD by 2022 from 1.05 billion USD in 2017, at a compound annual growth rate of 20.2%, there is an opportunity for this invention to compete with existing technologies.
Technology
The inventors introduced new elements to, and evaluated and optimized, most of the key steps involved in the cDNA library preparation method. Results for each step include:
1. 3’ dephosphorylation: The rSAP and Fast AP enzymes allow for greater dephosphorylation efficiency (>80%) than the PNK enzyme (~45%); their cost per reaction is also about 8–10 times cheaper than that of PNK.
2. 3’ adapter ligation: Using the molecular crowding agent polyethylene glycol to facilitate the ligation yield, and a higher ratio of RNA to adapter (from 1:1 to 1:10), greatly improved RNA ligation efficiency (from 65% to 81%, respectively).
3. Excess 3’ adapter digestion and clean up: Introducing two enzymes (a deadenylase and a 5’ to 3’ DNA exonuclease) to help with excess adapter digestion, followed by a column purification step to remove the residual adapters and clean up the sample, led to the digestion of the excess adapters by about 92%.
4. Reverse transcription of the ligated RNA-adapter: Modifying the reaction buffer to a Li+ containing reaction buffer (non-G-quadruplex stabilizing) led to no stalling band and more full-length band in the SSIII and protoscript reverse transcriptases. Using TGIRT with commercial buffer also alleviated the reverse transcriptase stalling. Of the 3 reverse transcriptases, the SSIII were the best, with 55.0% under the Li+ containing buffer.
5. Single-stranded DNA (ssDNA) ligation: At least 90% ssDNA ligation yield was achieved by optimizing several reaction conditions (e.g., temperature, PEG, ratio of cDNA to adapter); modifying a previously developed ssDNA ligation; and redesigning the adapter with a new sequence and base pairing strategy.
Advantages
- Compared with competing technologies, this invention requires similar or lower RNA input and has more efficient steps that will reduce the time and cost for cDNA library preparation.
Applications
- This invention can provide an efficient method for converting low-input RNA into a cDNA library.
- It can be used to produce a cDNA library ready for NGS.
- It can be applied to low-input RNA to determine the RNA expression or RNA structurome at a transcriptome-wide scale.
- It can be applied to low-input RNA for various NGS methods.