Involved Members: Dr. Leo Lai CHAN
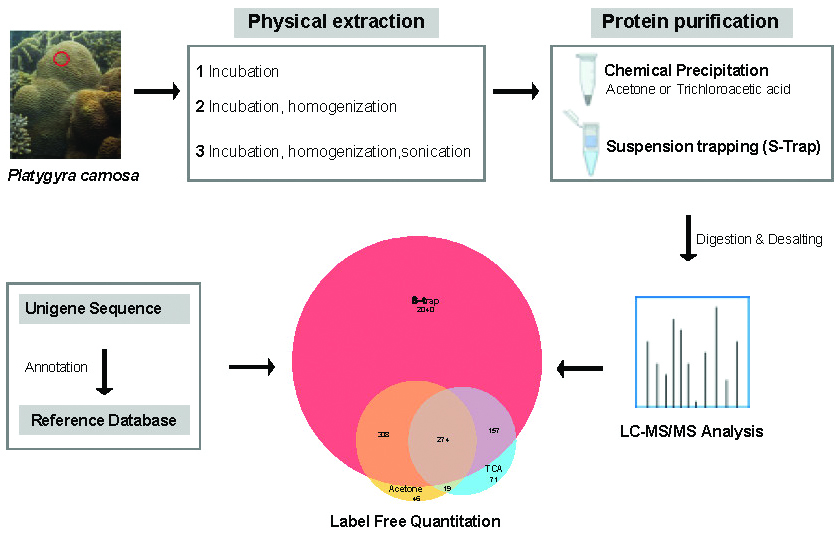
Stony corals form the foundation of coral reefs, which are of prominent ecological and economic significance. A robust workflow for investigating the coral proteome is essential in understanding coral biology. Here we investigated different preparative workflows and characterized the proteome of Platygyra carnosa, a common stony coral of the South China Sea. We found that a combination of bead homogenization with suspension trapping (S-Trap) preparation could yield more than 2700 proteins from coral samples. Annotation using a P. carnosa transcriptome database revealed that the majority of proteins were from the coral host cells. Label-free quantification and functional annotations indicated that a high proportion were involved in protein and redox homeostasis. Furthermore, the S-Trap method achieved good reproducibility in quantitative analysis. Although yielding a low symbiont: host ratio, the method is efficient in characterizing the coral host proteomic landscape, which provides a foundation to explore the molecular basis of the responses of coral host tissues to environmental stressors.
Reference:
Ma, H., Liao, H., Dellisanti, W., Sun, Y., Chan, L.L., & Zhang, L. (2021). Characterizing the host coral proteome of Platygyra carnosa using suspension trapping (S-Trap). Journal of Proteome Research, 20(3), 1783-1791.