An international research team co-led by Dr Yan Jian, Assistant Professor in the Department of Biomedical Sciences at City University of Hong Kong (CityU), has developed a high-throughput biological assay technique that provides valuable data for finding type-2 diabetes key biomarkers for diagnostics and treatment.
The findings, published in the prestigious scientific journal Nature under the title “Systematic analysis of binding of transcription factors to noncoding variants”, enables a systematic analysis of the impact of nearly 100,000 common genetic variants on the binding of transcription factors to DNA.
“Based on our findings, we believe that our high-throughput experimental method can be applied in the study of different genetic diseases, including colorectal cancer and prostate cancer. It can help dissect the mechanism of the genetic inheritance of the disease and find the biomarkers for clinical diagnosis, which will help us develop methods or strategies to prevent, detect or cure the diseases early,” said Dr Yan.
The other co-leaders of the project are Professor Bing Ren from the University of California San Diego and Professor Jussi Taipale from the University of Cambridge.
Genome-wide association studies (GWAS), which investigate the entire genome, constitute the most important strategy in finding genetic loci associated with complex genetic diseases. Researchers have found hundreds of thousands of genetic variants in association with human diseases and traits, but studies on the functions of these variants are still limited.
One of the variants’ function is to affect the binding of transcription factors to DNA. The transcription factors will then control the gene expression in cells, turning the specific gene expression “up” and “down”, modulating cellular functions.
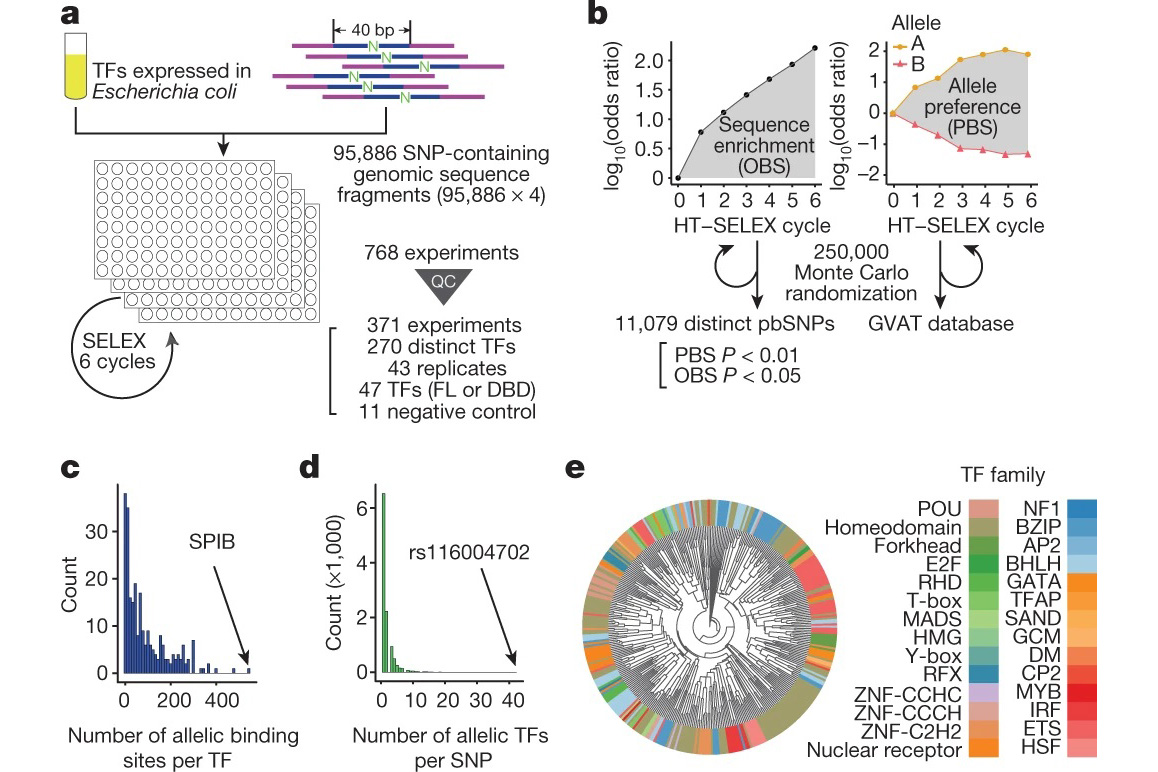
To systematically characterise the effects of genetic variants on the transcription factor binding, the team modified their previously developed experimental method an ultra-high-throughput multiplex protein-DNA binding assay HT-SELEX into an assay that can systematically and quantitatively evaluate the impact of a single nucleotide change on protein binding, termed “single-nucleotide polymorphism evaluation by systematic evolution of ligands by exponential enrichment” (SNP-SELEX). Then, they chose the genetic variants from the gene locations on the genome (called “gene loci”) that are known to be associated with the risk of type-2 diabetes as the object of analysis.
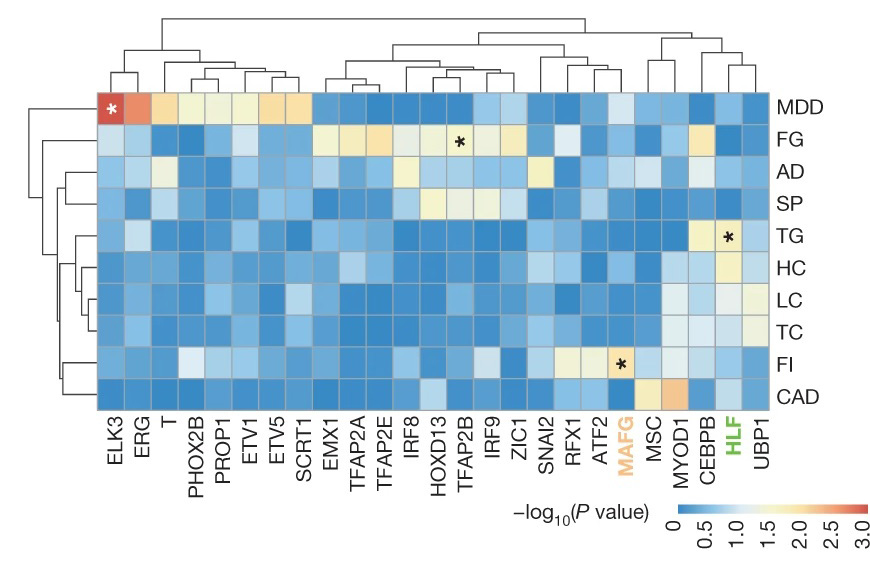
Utilising SNP-SELEX, the teams analysed the impact of 95,886 genetic variants on the binding of 270 distinct human transcription factors to DNA, demonstrating for example that the noncoding genetic variant SNP rs7118999 that increases the risk of type-2 diabetes can affect DNA binding with one of the transcription factors, and also that the resulting molecular mechanism regulates lipid levels in blood.
“This is a clear example of applying data generated by SNP-SELEX to help identify genetic variants in the inheritance of type 2 diabetes, which will help subsequent investigations into identifying diagnostic biomarkers and therapeutic targets,” said Dr Yan.
Dr Yan, Dr Qiu Yunjiang from the University of California San Diego and Dr André M. Ribeiro dos Santos from the Federal University of Pará in Brazil are the paper’s co-first authors. Dr Yan, Professor Ren and Professor Taipale are the co-corresponding authors. Other team members include researchers from Karolinska Institutet, University of Cambridge, University of California San Diego and CityU, coming from US, UK, Sweden, Brazil, mainland China and Hong Kong.